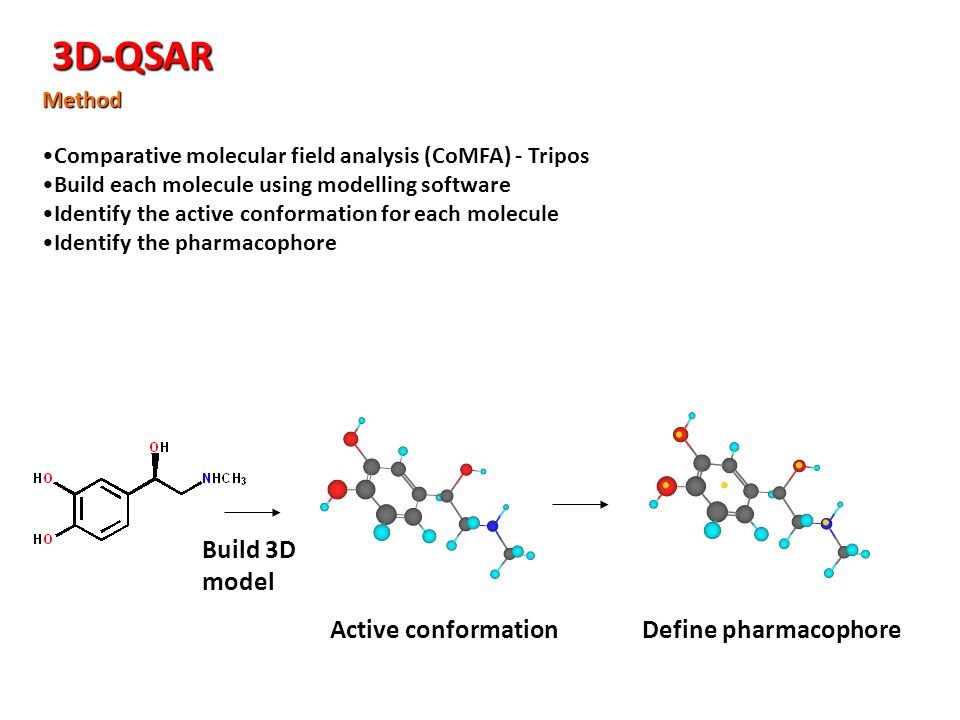
3d Qsar Software As A Service
Comparison of software for molecular mechanics modeling. From Wikipedia, the free encyclopedia. This is a list of computer programs that are predominantly used for. In this blog post I’m going to look at 3D-QSAR in Cresset’s software. I will share some of my experience on the best way to do it and, more importantly, what not. What is the best free software for QSAR and molecular docking? I have free online service or any other free online software for 3D QSAR??? Comparison of software for molecular mechanics modeling. From Wikipedia, the free encyclopedia. This is a list of computer programs that are predominantly used for.

Was presented by Dr Mark Mackey, Cresset at the Cresset European User Group Meeting 2015. Abstract 3D-QSAR based on molecular interaction potentials can provide a wealth of information about the exact molecular characteristics required for activity.
However, current techniques have a number of issues such as alignment noise, sampling errors and descriptor choice which can make it difficult to reliably produce effective models. We have presented in the past techniques for solving the sampling problem and shown that using accurate electrostatics combined with simple shape descriptors often gives meaningful models. However, there are still times when it is not possible to obtain a statistically valid linear regression model. One useful qualitative data analysis method that is being increasingly used is activity cliffs analysis. In this technique, pairs of compounds are located that are similar (in some sense), but have different activities.
Traditionally activity cliff analysis has used a 2D definition of similarity, but extension to 3D similarity metrics gives additional information that is very useful to locate the source of and reason for the activity differences. An extension of 3D activity cliff analysis is to mine the entire data set for corresponding cliffs and use this to build a model for activity. Analysis of the data set to locate activity cliffs locates the pairs of molecules with the highest information content. However, this needs to be tempered with an analysis of how likely it is that the molecules are aligned correctly, as only properly-aligned molecules contain any information. We apply Bayesian corrections to the activity cliff data to obtain a map of the electrostatic and shape characteristics that seem to locally correlate with improving activity.
Graphene Powerpoint (ppt) Template - Powerpoint Backgrounds On Graphene Templates more. The resulting model is semi-quantitative in that it attempts to describe the entire data set without building a linear regression model. This technique provides a valuable fall back to the computational chemist for information extraction from ligands in 3D.
Posted June 29th, 2015 by & filed under,. Was presented by Dr Bohdan Waszkowycz, Cancer Research at the Cresset European User Group Meeting 2015. The Cancer Research UK group at the Manchester Institute have been looking at ways to improve the selectivity of RET kinase inhibitors.
They used the 3D-QSAR models in and compared them with other 3D-QSAR methods. They concluded that building a successful and robust QSAR model takes time and patience, but Forge offers a user-friendly approach to building 3D-QSAR models. In recent years RET kinase has been implicated in lung cancer and thyroid cancer, so there is a large potential market for a compound that can inhibit it effectively.
There are a few known inhibitors used in the clinic, but they have not been designed specifically for the target but hit a range of kinases, so they have a lot of off-target toxicity. The group’s goal for this project was to identify novel inhibitors with improved potency and selectivity towards RET. The 3D-QSAR dataset Bohdan described the quinazoline dataset that they have been working on at Manchester.
They synthesized over 450 examples, with very varying properties, and then selected a subset of 128 compounds for this 3D-QSAR study to focus on the gatekeeper pocket of the kinase. They initially based their modeling on X-ray structures from the PDB, but later in the project they obtained their own in-house X-ray for a phenolic quinazoline analogue that was of interest because it showed a large boost in RET affinity and selectivity. Using Forge, they calculated the fields and difference maps for the simple phenol and a number of chemically diverse analogues to explore how the fields varied with substitution. They found that the phenol is important because it forms a couple of critical hydrogen bonds in the gatekeeper pocket. Why should we want to build a QSAR model? The goal of building a QSAR model is to find a correlation between chemical structure and biological activity.